This function performs non-hierarchical clustering based on dissimilarity using the Density-Based Spatial Clustering of Applications with Noise (DBSCAN) algorithm.
Usage
nhclu_dbscan(
dissimilarity,
index = names(dissimilarity)[3],
minPts = NULL,
eps = NULL,
plot = TRUE,
algorithm_in_output = TRUE,
verbose = TRUE,
...
)Arguments
- dissimilarity
The output object from
dissimilarity()orsimilarity_to_dissimilarity(), or adistobject. If adata.frameis used, the first two columns should represent pairs of sites (or any pair of nodes), and the subsequent column(s) should contain the dissimilarity indices.- index
The name or number of the dissimilarity column to use. By default, the third column name of
dissimilarityis used.- minPts
A
numericvector or a singlenumericvalue specifying theminPtsargument ofdbscan::dbscan().minPtsis the minimum number of points to form a dense region. By default, it is set to the natural logarithm of the number of sites indissimilarity. See Details for guidance on choosing this parameter.- eps
A
numericvector or a singlenumericvalue specifying theepsargument ofdbscan::dbscan().epsspecifies how similar points should be to each other to be considered part of a cluster. See Details for guidance on choosing this parameter.- plot
A
booleanindicating whether the k-nearest neighbor distance plot should be displayed.- algorithm_in_output
A
booleanindicating whether the original output of dbscan::dbscan should be included in the output. Defaults toTRUE(see Value).- verbose
A
booleanindicating whether to display progress messages. Set toFALSEto suppress these messages.- ...
Additional arguments to be passed to
dbscan()(see dbscan::dbscan).
Value
A list of class bioregion.clusters with five components:
name: A
characterstring containing the name of the algorithm.args: A
listof input arguments as provided by the user.inputs: A
listof characteristics of the clustering process.algorithm: A
listof all objects associated with the clustering procedure, such as original cluster objects (only ifalgorithm_in_output = TRUE).clusters: A
data.framecontaining the clustering results.
If algorithm_in_output = TRUE, the algorithm slot includes the output of
dbscan::dbscan.
Details
The DBSCAN (Density-Based Spatial Clustering of Applications with Noise)
algorithm clusters points based on the density of neighbors around each
data point. It requires two main arguments: minPts, the minimum number of
points to identify a core, and eps, the radius used to find neighbors.
Choosing minPts: This determines how many points are necessary to form a cluster. For example, what is the minimum number of sites expected in a bioregion? Choose a value sufficiently large for your dataset and expectations.
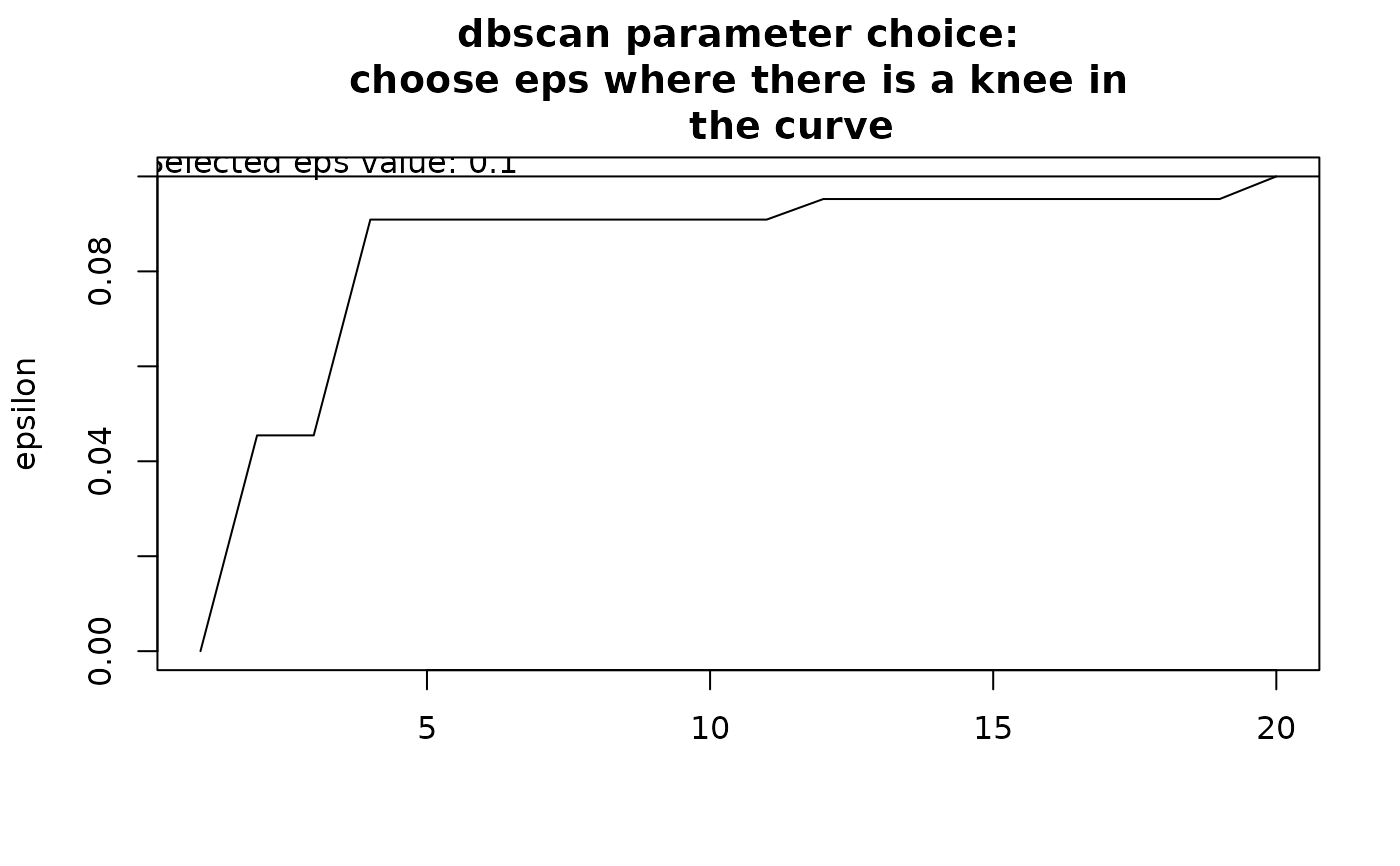
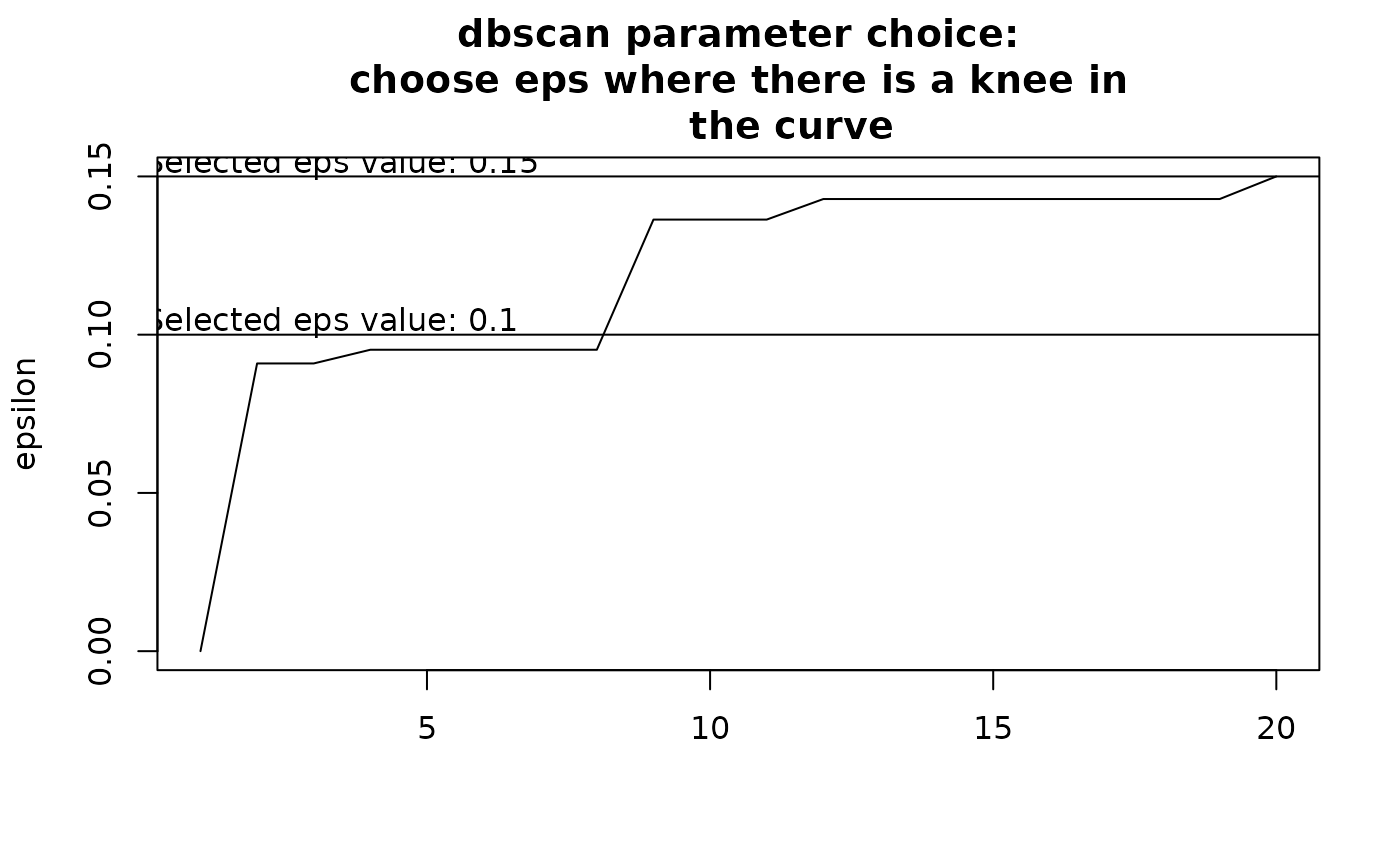
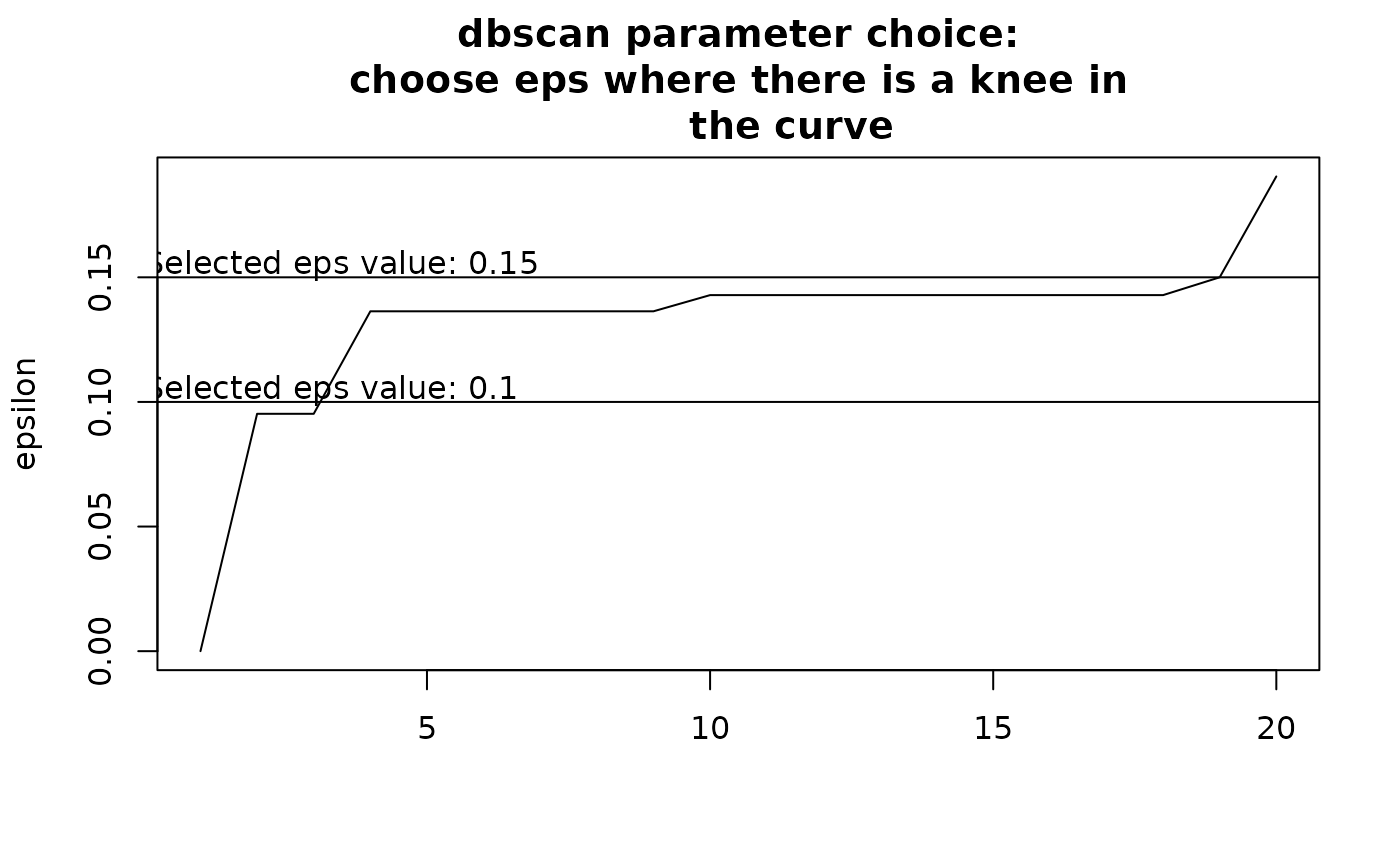
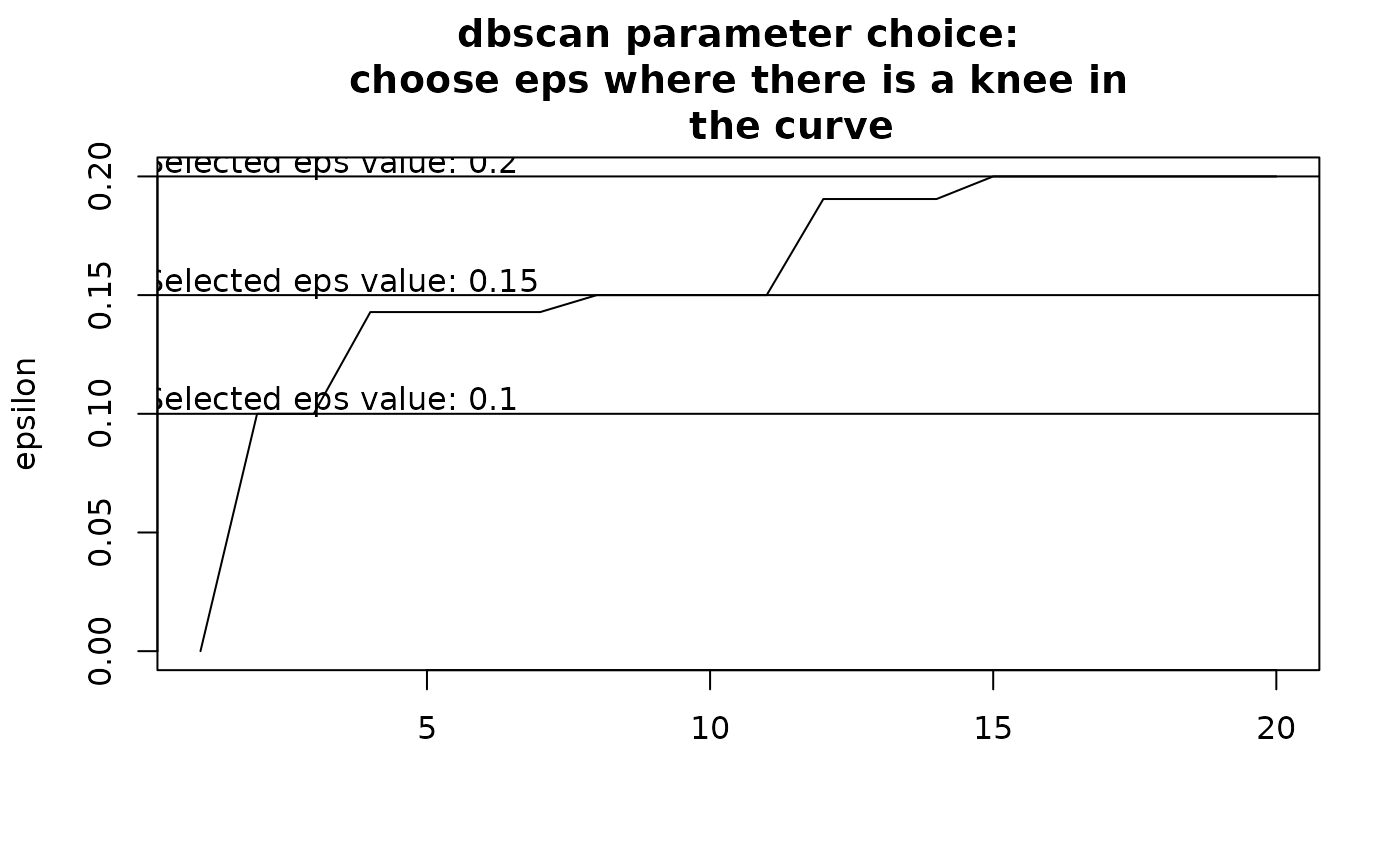
Choosing eps: This determines how similar sites should be to form a
cluster. If eps is too small, most points will be considered too distinct
and marked as noise. If eps is too large, clusters may merge. The value of
eps depends on minPts. It is recommended to choose eps by identifying
a knee in the k-nearest neighbor distance plot.
By default, the function attempts to find a knee in this curve
automatically, but the result is uncertain. Users should inspect the graph
and modify eps accordingly. To explore eps values, run the function
initially without defining eps, review the recommendations, and adjust
as needed based on clustering results.
References
Hahsler M, Piekenbrock M & Doran D (2019) Dbscan: Fast density-based clustering with R. Journal of Statistical Software, 91(1), 1–30.
See also
For more details illustrated with a practical example, see the vignette: https://biorgeo.github.io/bioregion/articles/a4_2_non_hierarchical_clustering.html.
Associated functions: nhclu_clara nhclu_clarans nhclu_kmeans nhclu_pam nhclu_affprop
Author
Boris Leroy (leroy.boris@gmail.com)
Pierre Denelle (pierre.denelle@gmail.com)
Maxime Lenormand (maxime.lenormand@inrae.fr)
Examples
comat <- matrix(sample(0:1000, size = 500, replace = TRUE, prob = 1/1:1001),
20, 25)
rownames(comat) <- paste0("Site",1:20)
colnames(comat) <- paste0("Species",1:25)
dissim <- dissimilarity(comat, metric = "all")
clust1 <- nhclu_dbscan(dissim, index = "Simpson")
#> Trying to find a knee in the curve to search for an optimal eps value...
#> NOTE: this automatic identification of the knee may not work properly
#> if the curve has knees and elbows. Please adjust eps manually by
#> inspecting the curve, identifying a knee as follows:
#>
#> /
#> curve /
#> ___________/ <- knee
#> elbow -> /
#> /
#> /
 clust2 <- nhclu_dbscan(dissim, index = "Simpson", eps = 0.2)
clust2 <- nhclu_dbscan(dissim, index = "Simpson", eps = 0.2)
 clust3 <- nhclu_dbscan(dissim, index = "Simpson", minPts = c(5, 10, 15, 20),
eps = c(.1, .15, .2, .25, .3))
clust3 <- nhclu_dbscan(dissim, index = "Simpson", minPts = c(5, 10, 15, 20),
eps = c(.1, .15, .2, .25, .3))