4.3 Network clustering
Pierre Denelle, Boris Leroy and Maxime Lenormand
2026-01-23
Source:vignettes/a4_3_network_clustering.Rmd
a4_3_network_clustering.RmdNetworks are objects made of nodes and links. The links connect the nodes. Networks can be bipartite, i.e. the nodes can be of two different types and one type can only be linked with the second one. Links can also be weighted.
Site-species matrices can be viewed as bipartite networks where sites and species are two different classes of nodes, and the occurrences of species within sites are the links.
The community detection algorithms try to identify the communities of nodes that interact more with each other than the network as a whole. Appling these algorithms to site-species matrices is therefore equivalent to the more classic bioregionalization methods.
The optimal community partition of a network can be found by searching for the partition that maximizes a metric called modularity. Maximising the modularity is a computationally hard problem requiring the use of algorithms.
Network theory offers plenty of such algorithms to classify nodes
that are more connected than expected randomly based or not on the
modularity. With the bioregion R package, we cover the main
network algorithms to identify bioregions in site-species matrices. All
functions relying on network algorithms start with the prefix
netclu_.
Network clustering takes place on the left-hand size part of the
bioregion conceptual diagram:
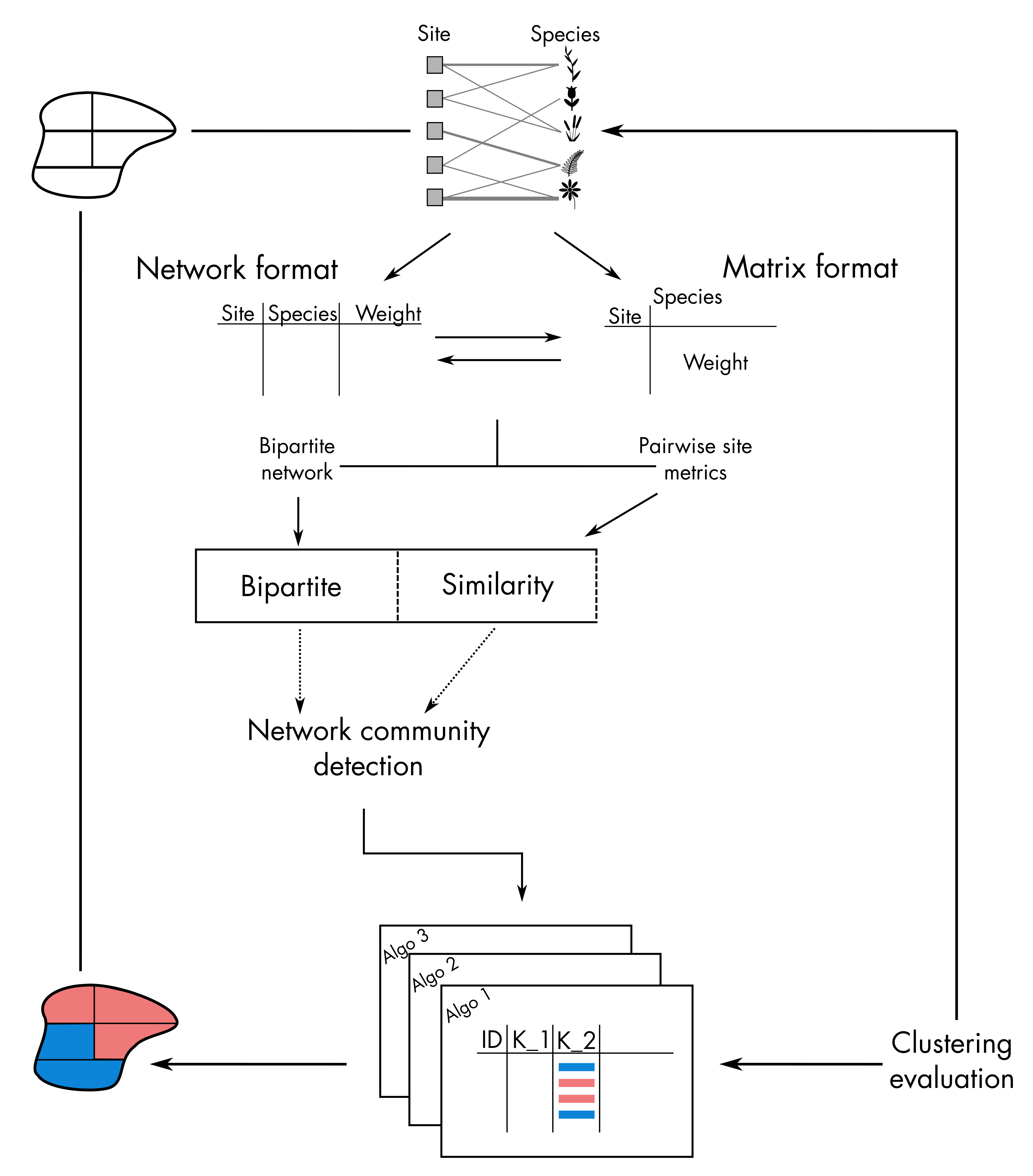
In total, we have 9 functions, which can be classified like this:
- Functions based on binary files
-
netclu_infomap
-
netclu_oslom
- netclu_louvain
- Functions based on the igraph package
* netclu_louvain
* netclu_greedy
* netclu_labelprop
* netclu_leadingeigen
* netclu_walktrap
- Function based on the bipartite package
* netclu_beckett
This vignette aims at briefly explaining how each algorithm and its
associated bioregion function works. For this purpose, we
use the European freshwater fish dataset that comes with
bioregion.
1 Introduction
1.1 Input data
All network algorithms work with the network format, i.e. a
data.frame with 3 columns: sites, species and the abundance
of a given species in a given site. This type of object can be obtained
from a site x species matrix through the use of
mat_to_net().
In this vignette, we directly load the network format for the distribution of fish in European basins.
library("bioregion")
data("fishdf")Some network algorithms work with the similarity matrix between each pair of sites.
data("fishmat")
fish_simil <- similarity(fishmat, metric = "Simpson")1.2 Main arguments
Each of the algorithms presented here has some specific parameters that can be tweaked but some arguments are common for all the functions.
Among these common arguments are the following:
* weight a boolean indicating if the weights should be
considered
* index name or number of the column to use as weight. By
default, the third column name of the network data.frame is used
* site_col name or number for the column of site nodes
(i.e. primary nodes).
* species_col = name or number for the column of species
nodes (i.e. feature nodes)
* return_node_type a character indicating what types of
nodes (“sites”, “species” or “both”) should be returned in the output
(keep_nodes_type=“both” by default).
* algorithm_in_output a boolean indicating if the original
output of communities should be returned in the output (see Value).
For the three algorithms relying on executable binary files, the
following arguments are needed:
* delete_temp a boolean indicating if the temporary folder
should be removed
* path_temp a character indicating the path to the
temporary folder
* binpath a character indicating the path to the bin
folder
2. Binary files
install_binaries(binpath = "tempdir", infomap_version = c("2.1.0", "2.6.0"))2.1 Infomap
Rosvall & Bergstrom (2008)
-
nbmodpenalize solutions the more they differ from this number (0 by default for no preferred number of modules).
-
markovtimescales link flow to change the cost of moving between modules, - higher values results in fewer modules (default is 1).
-
seedfor the random number generator (NULL for random by default)
-
numtrialsthe number of trials before picking up the best solution.
-
twolevela boolean indicating if the algorithm should optimize a two-level partition of the network (default is multi-level).
-
show_hierarchya boolean specifying if the hierarchy of community should be identifiable in the outputs (FALSE by default).
set.seed(1)
ex_infomap <- netclu_infomap(fish_simil,
weight = TRUE,
index = names(fish_simil)[3],
nbmod = 0,
markovtime = 1,
seed = NULL,
numtrials = 1,
twolevel = FALSE,
show_hierarchy = FALSE,
directed = FALSE,
bipartite_version = FALSE,
bipartite = FALSE,
site_col = 1,
species_col = 2,
return_node_type = "both",
version = "2.6.0",
binpath = "tempdir",
path_temp = "infomap_temp",
delete_temp = TRUE)
table(ex_infomap$clusters$K_5)##
## 1 2 3 4 5
## 290 14 23 9 22.2 OSLOM
OSLOM stands for Order Statistics Local Optimization Method.
Similarity-based algorithm.
Lancichinetti et al. (2011)
reassign a string indicating if the nodes belonging to
several community should be reassign and what method should be used (see
Note).
* r the number of runs for the first hierarchical level (10
by default).
* hr the number of runs for the higher hierarchical level
(50 by default, 0 if you are not interested in hierarchies).
* seed for the random number generator (NULL for random by
default).
* t the p-value, the default value is 0.10, increase this
value you to get more modules.
* cp kind of resolution parameter used to decide between
taking some modules or their union (default value is 0.5, bigger value
leads to bigger clusters).
set.seed(1)
ex_oslom <- netclu_oslom(fish_simil,
weight = TRUE,
index = names(fish_simil)[3],
reassign = "no",
r = 10,
hr = 50,
seed = NULL,
t = 0.1,
cp = 0.5,
directed = FALSE,
bipartite = FALSE,
site_col = 1,
species_col = 2,
return_node_type = "both",
binpath = "tempdir",
path_temp = "oslom_temp",
delete_temp = TRUE)
table(ex_oslom$clusters$K_338)2.3 Louvain
Blondel et al. (2008)
-
qthe quality function used to compute partition of the graph (modularity is chosen by default, see Details). -
cthe parameter for the Owsinski-Zadrozny quality function (between 0 and 1, 0.5 is chosen by default) -
kthe kappa_min value for the Shi-Malik quality function (it must be > 0, 1 is chosen by default)
set.seed(1)
ex_louvain <- netclu_louvain(fishdf,
weight = TRUE,
index = names(fishdf)[3],
lang = "cpp",
q = 0,
c = 0.5,
k = 1,
bipartite = FALSE,
site_col = 1,
species_col = 2,
return_node_type = "both",
binpath = "tempdir",
path_temp = "louvain_temp",
delete_temp = TRUE,
algorithm_in_output = TRUE)
table(ex_louvain$clusters$K_23)## < table of extent 0 >3. Functions from the igraph package
3.1 Fastgreedy
Clauset et al. (2004)
set.seed(1)
ex_greedy <- netclu_greedy(fishdf,
weight = TRUE,
index = names(fishdf)[3],
bipartite = FALSE,
site_col = 1,
species_col = 2,
return_node_type = "both",
algorithm_in_output = TRUE)
table(ex_greedy$clusters$K_5)##
## 1 2 3 4 5
## 138 61 144 132 583.2 Label propagation
Raghavan et al. (2007)
set.seed(1)
ex_labelprop <- netclu_labelprop(fishdf,
weight = TRUE,
index = names(fishdf)[3],
bipartite = FALSE,
site_col = 1,
species_col = 2,
return_node_type = "both",
algorithm_in_output = TRUE)
table(ex_labelprop$clusters$K_11)## < table of extent 0 >3.3 Leiden algorithm
Traag et al. (2019)
set.seed(1)
ex_leiden <- netclu_leiden(fishdf,
weight = TRUE,
index = names(fishdf)[3],
objective_function = "CPM",
resolution_parameter = 1,
beta = 0.01,
n_iterations = 2,
vertex_weights = NULL,
bipartite = TRUE,
site_col = 1,
species_col = 2,
return_node_type = "both",
algorithm_in_output = TRUE)
length(unique(ex_leiden$clusters$K_505))## [1] 03.4 Leading eigenvector
Newman (2006)
set.seed(1)
ex_leadingeigen <- netclu_leadingeigen(fishdf,
weight = TRUE,
index = names(fishdf)[3],
bipartite = FALSE,
site_col = 1,
species_col = 2,
return_node_type = "both",
algorithm_in_output = TRUE)
table(ex_leadingeigen$clusters$K_17)## < table of extent 0 >3.5 Walktrap
Pons & Latapy (2005)
set.seed(1)
ex_walktrap <- netclu_walktrap(fishdf,
weight = TRUE,
index = names(fishdf)[3],
steps = 4,
bipartite = FALSE,
site_col = 1,
species_col = 2,
return_node_type = "both",
algorithm_in_output = TRUE)
table(ex_walktrap$clusters$K_14)##
## 1 10 11 12 13 14 2 3 4 5 6 7 8 9
## 11 47 10 2 6 17 84 5 37 16 5 270 4 194 Function from the bipartite package
4.1 Beckett
Update of the QuanBiMo algorithm developed by Dormann & Strauss (2014).
Beckett (2016)
set.seed(1)
ex_beckett <- netclu_beckett(fishdf,
weight = TRUE,
index = names(fishdf)[3],
site_col = 1,
species_col = 2,
return_node_type = "both",
forceLPA = TRUE,
algorithm_in_output = TRUE)
ex_beckett$clusters$K_23## [1] "4" "1" "2" "3" "4" "4" "3" "3" "16" "13" "1" "1" "8" "1" "16"
## [16] "5" "4" "3" "4" "8" "4" "1" "4" "4" "5" "4" "4" "1" "13" "1"
## [31] "4" "3" "1" "3" "4" "4" "4" "4" "3" "3" "4" "3" "1" "5" "3"
## [46] "3" "6" "4" "3" "4" "1" "4" "1" "4" "4" "4" "7" "8" "4" "9"
## [61] "4" "4" "4" "4" "10" "10" "1" "4" "5" "1" "5" "4" "8" "8" "4"
## [76] "1" "3" "1" "16" "13" "12" "1" "22" "4" "3" "1" "3" "1" "13" "4"
## [91] "4" "8" "4" "8" "15" "15" "15" "15" "4" "1" "8" "4" "1" "8" "4"
## [106] "8" "4" "1" "1" "4" "1" "15" "1" "1" "5" "1" "16" "8" "1" "1"
## [121] "1" "13" "1" "8" "1" "8" "17" "18" "1" "1" "1" "8" "1" "19" "1"
## [136] "1" "8" "4" "4" "4" "4" "5" "4" "1" "1" "1" "4" "4" "5" "5"
## [151] "3" "8" "1" "11" "1" "4" "1" "1" "1" "1" "13" "16" "4" "1" "1"
## [166] "1" "1" "1" "1" "3" "8" "5" "5" "20" "5" "4" "3" "1" "4" "1"
## [181] "4" "1" "6" "1" "13" "3" "8" "8" "4" "3" "1" "8" "1" "4" "8"
## [196] "1" "3" "4" "1" "4" "8" "4" "1" "1" "4" "1" "16" "1" "4" "8"
## [211] "1" "3" "15" "1" "1" "16" "13" "1" "3" "1" "1" "1" "3" "1" "21"
## [226] "1" "4" "1" "1" "1" "1" "1" "4" "1" "1" "1" "8" "4" "4" "4"
## [241] "1" "22" "1" "1" "1" "4" "1" "5" "1" "4" "1" "1" "1" "8" "15"
## [256] "4" "1" "4" "4" "1" "4" "4" "4" "8" "3" "4" "8" "3" "4" "1"
## [271] "1" "13" "1" "4" "22" "1" "1" "3" "3" "5" "5" "4" "4" "1" "4"
## [286] "4" "4" "4" "3" "11" "4" "3" "4" "1" "1" "1" "1" "8" "4" "4"
## [301] "4" "4" "3" "1" "5" "1" "1" "4" "5" "8" "1" "1" "1" "1" "1"
## [316] "5" "1" "4" "1" "1" "14" "10" "8" "4" "1" "4" "4" "4" "4" "3"
## [331] "1" "1" "5" "4" "8" "8" "4" "23" "4" "4" "4" "8" "8" "1" "4"
## [346] "4" "4" "13" "1" "4" "4" "1" "16" "16" "2" "16" "2" "2" "2" "16"
## [361] "2" "16" "3" "4" "3" "3" "3" "3" "3" "3" "3" "3" "4" "5" "4"
## [376] "4" "3" "3" "12" "16" "13" "13" "13" "13" "13" "13" "13" "13" "13" "13"
## [391] "13" "13" "13" "8" "8" "16" "16" "5" "20" "15" "5" "5" "4" "3" "3"
## [406] "8" "15" "5" "5" "5" "5" "5" "5" "13" "8" "13" "9" "3" "3" "3"
## [421] "5" "6" "19" "6" "6" "7" "7" "7" "7" "7" "8" "7" "7" "7" "7"
## [436] "21" "21" "7" "21" "7" "7" "7" "7" "7" "7" "8" "10" "8" "10" "9"
## [451] "9" "9" "9" "10" "9" "9" "8" "10" "22" "9" "9" "9" "10" "9" "8"
## [466] "10" "9" "9" "4" "10" "10" "8" "10" "10" "15" "5" "5" "11" "11" "15"
## [481] "13" "13" "22" "22" "13" "22" "12" "12" "22" "13" "13" "15" "15" "14" "15"
## [496] "15" "15" "15" "15" "5" "16" "17" "18" "18" "18" "18" "23" "19" "19" "13"
## [511] "20" "13" "22" "13" "13" "13" "3" "21" "21" "21" "21" "21" "21" "4" "22"
## [526] "22" "5" "22" "22" "22" "5" "10" "23"